




Python-based software for generalized object instance segmentation from (cryo-)electron microscopy micrographs/tomograms. The software package is built on a general workflow where predicted semantic segmentation is used for instance segmentation of 2D/3D images.
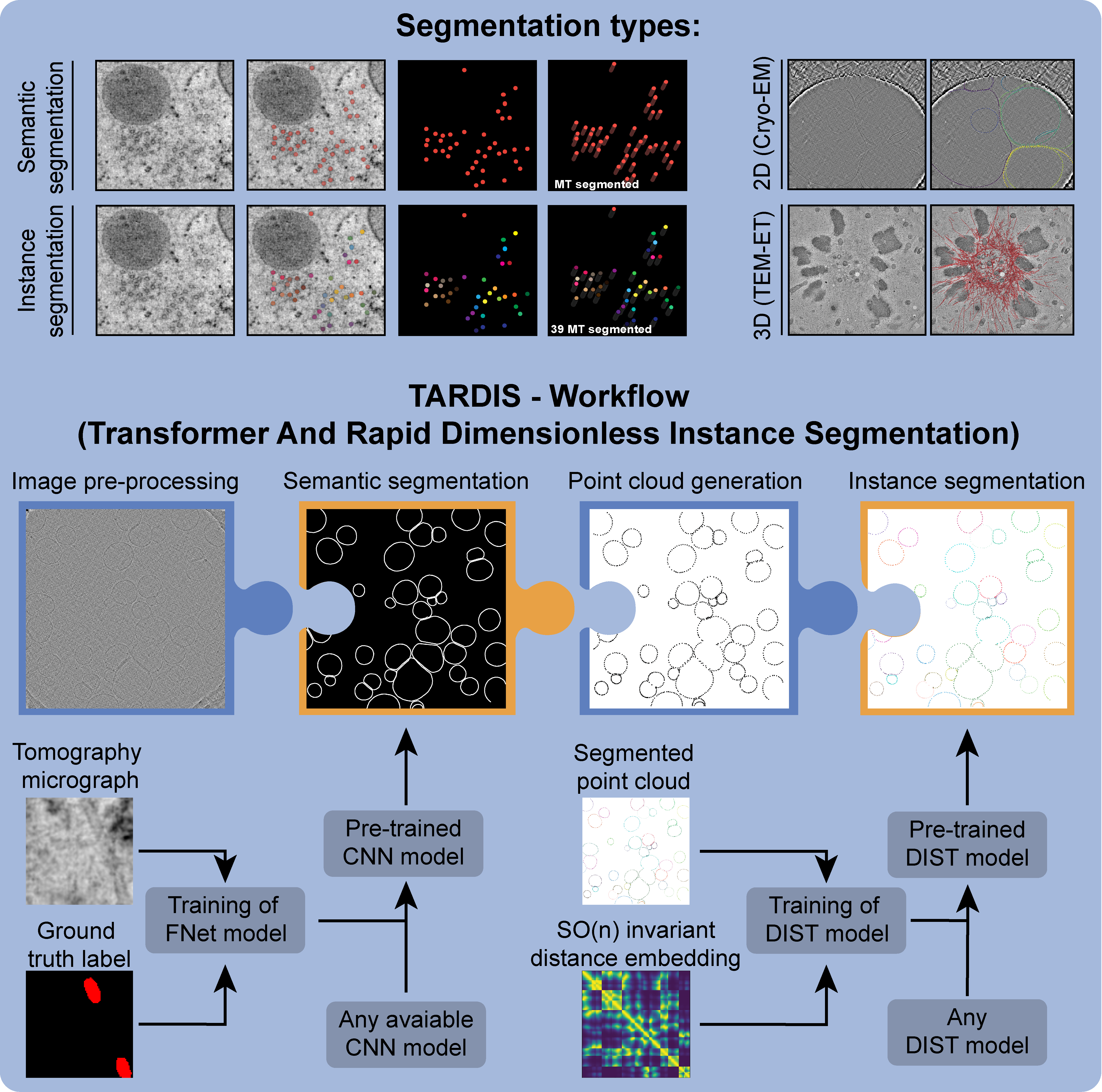
Features
- Robust and high-throughput semantic/instance segmentation of all microtubules:
Supported file formats: [.tif, .mrc, .rec, .am]
Supported modality: [ET, Cryo-ET]
Supported Å resolution: [any best results in 1-40 Å range]
2D micrograph modality microtubule segmentation will come soon!
- Robust and high-throughput semantic/instance segmentation of membranes:
Supported file formats: [.tif, .mrc, .rec, .am]
Supported modality: [EM, ET, Cryo-EM, Cryo-ET]
Supported Å resolution: [all]
High-throughput semantic/instance segmentation of actin [Beta]
Fully automatic segmentation solution!
Cloud computing [Coming soon]
Citation
Kiewisz R. et.al. 2024. Accurate and fast segmentation of filaments and membranes in micrographs and tomograms with TARDIS.
DOI [Microscopy and Microanalysis]
Kiewisz R., Fabig G., Müller-Reichert T. Bepler T. 2023. Automated Segmentation of 3D Cytoskeletal Filaments from Electron Micrographs with TARDIS. Microscopy and Microanalysis 29(Supplement_1):970-972.
Link: NeurIPS 2022 MLSB Workshop
Kiewisz R., Bepler T. 2022. Membrane and microtubule rapid instance segmentation with dimensionless instance segmentation by learning graph representations of point clouds. Neurips 2022 - Machine Learning for Structural Biology Workshop.
What’s new?
- TARDIS-em v0.3.10 (2025-01-07):
Models update
Progress with estimated ETA time
Small bugfixes
Quick Start
For more examples and advanced usage please find more details in our Documentation
Install TARDIS-em:
Install pytorch with GPU support as per Pytorch official website: https://pytorch.org/get-started/locally/
pip install tardis-em
Verifies installation:
tardis
Optional Napari plugin installation
pip install napari-tardis-em
Filaments Prediction
3D Actin prediction
Full tutorial: 3D Actin Prediction
Usage:
recommended usage: tardis_actin [-dir path/to/folder/with/input/tomogram]
advance usage: tardis_actin [-dir str] [-out str] [-ps int] [-ct float] [-dt float]
[-pv int] [-px float] ...
2D Microtubule prediction
TBD
3D Microtubule prediction
Full tutorial: 3D Microtubules Prediction
Example:
Data source: Dr. Gunar Fabig and Prof. Dr. Thomas Müller-Reichert, TU Dresden
Usage:
recommended usage: tardis_mt [-dir path/to/folder/with/input/tomogram]
advance usage: tardis_mt [-dir str] [-out str] [-ps int] [-ct float] [-dt float]
[-pv int] [-px float] ...
TIRF Microtubule prediction
Full tutorial: TIRF Microtubules Prediction
Example:
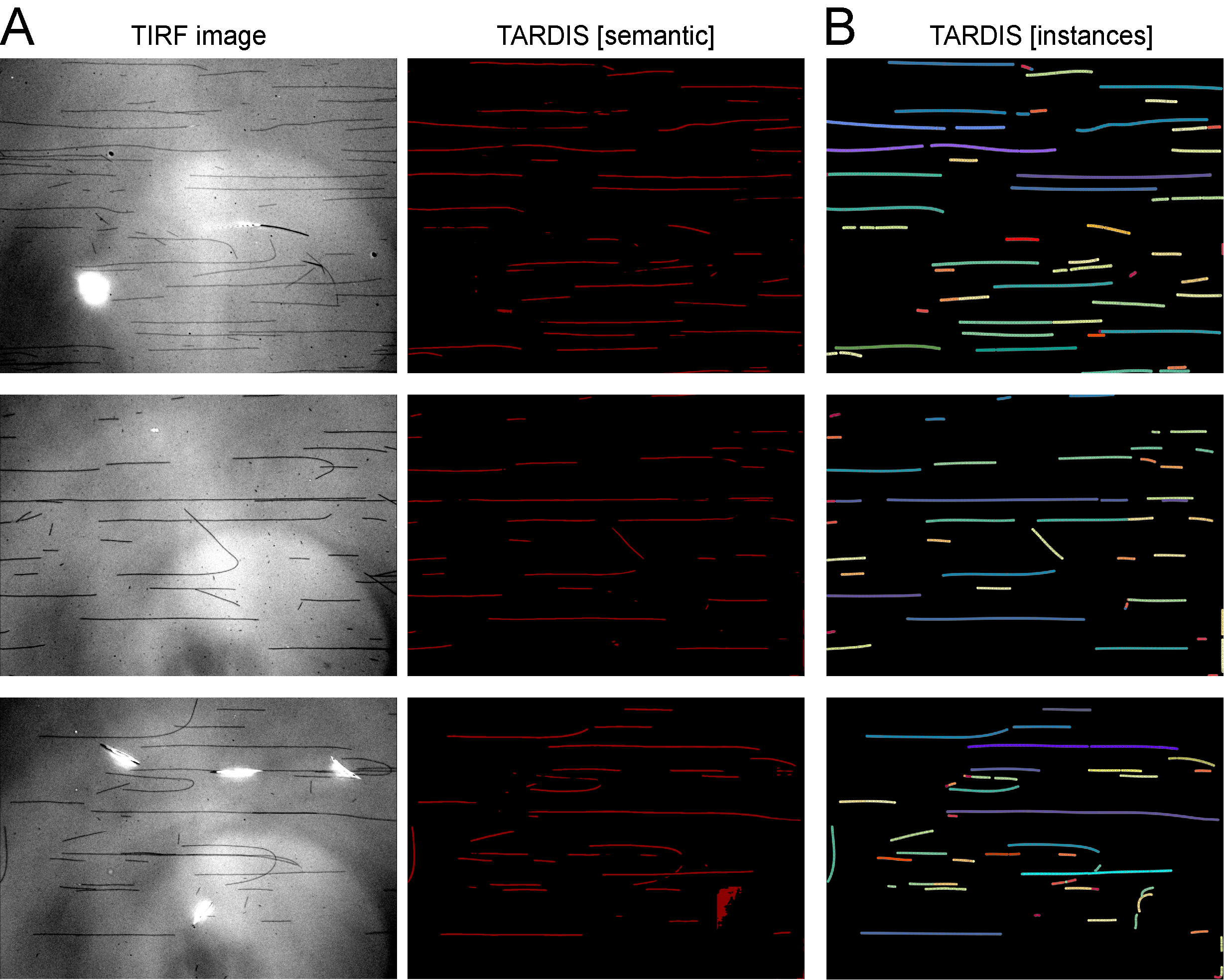
Data source: RNDr. Cyril Bařinka, Ph.D, Biocev
Usage:
recommended usage: tardis_mt_tirf [-dir path/to/folder/with/input/data]
advance usage: tardis_mt [-dir str] [-out str] [-ps int] [-ct float] [-dt float]
[-pv int] ...
Membrane Prediction
2D prediction
Full tutorial: 2D Membrane Prediction
Example:

Data source: Dr. Victor Kostyuchenko and Prof. Dr. Shee-Mei Lok, DUKE-NUS Medical School Singapore
Usage:
recommended usage: tardis_mem2d [-dir path/to/folder/with/input/tomogram] -out mrc_csv
advance usage: tardis_mem [-dir str] [-out str] [-ps int] ...
3D prediction
Full tutorial: 3D Membrane Prediction
Example:
Data source: EMPIRE-10236, DOI: 10.1038/s41586-019-1089-3
Usage:
recommended usage: tardis_mem [-dir path/to/folder/with/input/tomogram] -out mrc_csv
advance usage: tardis_mem [-dir str] [-out str] [-ps int] ...